Perform spatial error estimation and variable importance assessment
Source:R/sperrorest.R
sperrorest.Rdsperrorest is a flexible interface for multiple types of parallelized spatial and non-spatial cross-validation and bootstrap error estimation and parallelized permutation-based assessment of spatial variable importance.
sperrorest(
formula,
data,
coords = c("x", "y"),
model_fun,
model_args = list(),
pred_fun = NULL,
pred_args = list(),
smp_fun = partition_cv,
smp_args = list(),
train_fun = NULL,
train_param = NULL,
test_fun = NULL,
test_param = NULL,
err_fun = err_default,
imp_variables = NULL,
imp_permutations = 1000,
imp_sample_from = c("test", "train", "all"),
importance = !is.null(imp_variables),
distance = FALSE,
do_gc = 1,
progress = "all",
benchmark = FALSE,
mode_rep = c("future", "sequential", "loop"),
mode_fold = c("sequential", "future", "loop"),
verbose = 0
)Arguments
- formula
A formula specifying the variables used by the
model. Only simple formulas without interactions or nonlinear terms should be used, e.g.y~x1+x2+x3but noty~x1*x2+log(x3). Formulas involving interaction and nonlinear terms may possibly work for error estimation but not for variable importance assessment, but should be used with caution. The formulay~...is not supported, buty~1(i.e. no predictors) is.- data
a
data.framewith predictor and response variables. Training and test samples will be drawn from this data set bytrain_funandtest_fun, respectively.- coords
vector of length 2 defining the variables in
datathat contain the x and y coordinates of sample locations.- model_fun
Function that fits a predictive model, such as
glmorrpart. The function must accept at least two arguments, the first one being a formula and the second a data.frame with the learning sample.- model_args
Arguments to be passed to
model_fun(in addition to theformulaanddataargument, which are provided by sperrorest)- pred_fun
Prediction function for a fitted model object created by
model. Must accept at least two arguments: the fittedobjectand adata.framenewdatawith data on which to predict the outcome.- pred_args
(optional) Arguments to
pred_fun(in addition to the fitted model object and thenewdataargument, which are provided by sperrorest).- smp_fun
A function for sampling training and test sets from
data. E.g. partition_kmeans for spatial cross-validation using spatial k-means clustering.- smp_args
(optional) Arguments to be passed to
smp_fun.- train_fun
(optional) A function for resampling or subsampling the training sample in order to achieve, e.g., uniform sample sizes on all training sets, or maintaining a certain ratio of positives and negatives in training sets. E.g. resample_uniform or resample_strat_uniform.
- train_param
(optional) Arguments to be passed to
resample_fun.- test_fun
(optional) Like
train_funbut for the test set.- test_param
(optional) Arguments to be passed to
test_fun.- err_fun
A function that calculates selected error measures from the known responses in
dataand the model predictions delivered bypred_fun. E.g. err_default (the default).- imp_variables
(optional; used if
importance = TRUE). Variables for which permutation-based variable importance assessment is performed. Ifimportance = TRUEandimp_variables==NULL, all variables informulawill be used.- imp_permutations
(optional; used if
importance = TRUE). Number of permutations used for variable importance assessment.- imp_sample_from
(default:
"test"): specified if the permuted feature values should be taken from the test set, the training set (a rather unlikely choice), or the entire sample ("all"). The latter is useful in leave-one-out resampling situations where the test set is simply too small to perform any kind of resampling. In any case importances are always estimates on the test set. (Note that resampling with replacement is used if the test set is larger than the set from which the permuted values are to be taken.)- importance
logical (default:
FALSE): perform permutation-based variable importance assessment?- distance
logical (default:
FALSE): ifTRUE, calculate mean nearest-neighbour distances from test samples to training samples using add.distance.represampling.- do_gc
numeric (default: 1): defines frequency of memory garbage collection by calling gc; if
< 1, no garbage collection; if>= 1, run a gc after each repetition; if>= 2, after each fold.- progress
character (default:
all): Whether to show progress information (if possible). Default shows repetition, fold and (if enabled) variable importance progress. Set to"rep"for repetition information only orFALSEfor no progress information.- benchmark
(optional) logical (default:
FALSE): ifTRUE, perform benchmarking and returnsperrorestbenchmarkobject.- mode_rep, mode_fold
character (default:
"future"and"sequential", respectively): specifies whether to parallelize the execution at the repetition level, at the fold level, or not at all. Parallel execution usesfuture.apply::future_lapply()(see details below). It is only possible to parallelize at the repetition level or at the fold level. The"loop"option uses aforloop instead of anlappyfunction; this option is for debugging purposes.- verbose
Controls the amount of information printed while processing. Defaults to 0 (no output).
Value
A list (object of class sperrorest) with (up to) six components:
error_rep:
sperrorestreperrorcontaining predictive performances at the repetition levelerror_fold:
sperroresterrorobject containing predictive performances at the fold levelrepresampling: represampling object
importance:
sperrorestimportanceobject containing permutation-based variable importances at the fold levelbenchmark:
sperrorestbenchmarkobject containing information on the system the code is running on, starting and finishing times, number of available CPU cores and runtime performancepackage_version:
sperrorestpackageversionobject containing information about the sperrorest package version
Details
Custom predict functions passed to pred_fun, which consist of
multiple child functions, must be defined in one function.
Parallelization
Running in parallel is supported via package future.
Have a look at vignette("future-1-overview", package = "future").
In short: Choose a backend and specify the number of workers, then call
sperrorest() as usual. Example:
future::plan(future.callr::callr, workers = 2)
sperrorest()Parallelization at the repetition is recommended when using repeated cross-validation. If the 'granularity' of parallelized function calls is too fine, the overall runtime will be very poor since the overhead for passing arguments and handling environments becomes too large. Use fold-level parallelization only when the processing time of individual folds is very large and the number of repetitions is small or equals 1.
Note that nested calls to future are not possible.
Therefore a sequential sperrorest call should be used for
hyperparameter tuning in a nested cross-validation.
References
Brenning, A. 2012. Spatial cross-validation and bootstrap for the assessment of prediction rules in remote sensing: the R package 'sperrorest'. 2012 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), 23-27 July 2012, p. 5372-5375. https://ieeexplore.ieee.org/document/6352393
Brenning, A. 2005. Spatial prediction models for landslide hazards: review, comparison and evaluation. Natural Hazards and Earth System Sciences, 5(6), 853-862. doi:10.5194/nhess-5-853-2005
Brenning, A., S. Long & P. Fieguth. 2012. Detecting rock glacier flow structures using Gabor filters and IKONOS imagery. Remote Sensing of Environment, 125, 227-237. doi:10.1016/j.rse.2012.07.005
Russ, G. & A. Brenning. 2010a. Data mining in precision agriculture: Management of spatial information. In 13th International Conference on Information Processing and Management of Uncertainty, IPMU 2010; Dortmund; 28 June - 2 July 2010. Lecture Notes in Computer Science, 6178 LNAI: 350-359.
Russ, G. & A. Brenning. 2010b. Spatial variable importance assessment for yield prediction in Precision Agriculture. In Advances in Intelligent Data Analysis IX, Proceedings, 9th International Symposium, IDA 2010, Tucson, AZ, USA, 19-21 May 2010. Lecture Notes in Computer Science, 6065 LNCS: 184-195.
Examples
## ------------------------------------------------------------
## Classification tree example using non-spatial partitioning
## ------------------------------------------------------------
# Muenchow et al. (2012), see ?ecuador
fo <- slides ~ dem + slope + hcurv + vcurv + log.carea + cslope
library(rpart)
mypred_part <- function(object, newdata) predict(object, newdata)[, 2]
ctrl <- rpart.control(cp = 0.005) # show the effects of overfitting
# show the effects of overfitting
fit <- rpart(fo, data = ecuador, control = ctrl)
### Non-spatial cross-validation:
mypred_part <- function(object, newdata) predict(object, newdata)[, 2]
nsp_res <- sperrorest(
data = ecuador, formula = fo,
model_fun = rpart,
model_args = list(control = ctrl),
pred_fun = mypred_part,
progress = TRUE,
smp_fun = partition_cv,
smp_args = list(repetition = 1:2, nfold = 3)
)
#> Thu Jan 30 04:22:55 2025 Repetition 1
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 1
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 2
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 3
#> Thu Jan 30 04:22:55 2025 Repetition 2
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 1
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 2
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 3
summary(nsp_res$error_rep)
#> mean sd median IQR
#> train_auroc 0.8413531 0.0002190341 0.8413531 0.0001548805
#> train_error 0.1917443 0.0018831073 0.1917443 0.0013315579
#> train_accuracy 0.8082557 0.0018831073 0.8082557 0.0013315579
#> train_sensitivity 0.8855000 0.0120208153 0.8855000 0.0085000000
#> train_specificity 0.6543825 0.0295801642 0.6543825 0.0209163347
#> train_fpr70 0.2051793 0.0140857925 0.2051793 0.0099601594
#> train_fpr80 0.2559761 0.0042257377 0.2559761 0.0029880478
#> train_fpr90 0.3954183 0.0042257377 0.3954183 0.0029880478
#> train_tpr80 0.6990000 0.0268700577 0.6990000 0.0190000000
#> train_tpr90 0.4380000 0.0212132034 0.4380000 0.0150000000
#> train_tpr95 0.2870000 0.0141421356 0.2870000 0.0100000000
#> train_events 1000.0000000 0.0000000000 1000.0000000 0.0000000000
#> train_count 1502.0000000 0.0000000000 1502.0000000 0.0000000000
#> test_auroc 0.5913347 0.0145196349 0.5913347 0.0102669323
#> test_error 0.3741678 0.0000000000 0.3741678 0.0000000000
#> test_accuracy 0.6258322 0.0000000000 0.6258322 0.0000000000
#> test_sensitivity 0.7420000 0.0169705627 0.7420000 0.0120000000
#> test_specificity 0.3944223 0.0338059019 0.3944223 0.0239043825
#> test_fpr70 0.5517928 0.0084514755 0.5517928 0.0059760956
#> test_fpr80 0.6812749 0.0225372679 0.6812749 0.0159362550
#> test_fpr90 0.8705179 0.0366230604 0.8705179 0.0258964143
#> test_tpr80 0.2720000 0.0678822510 0.2720000 0.0480000000
#> test_tpr90 0.1590000 0.0183847763 0.1590000 0.0130000000
#> test_tpr95 0.0920000 0.0056568542 0.0920000 0.0040000000
#> test_events 500.0000000 0.0000000000 500.0000000 0.0000000000
#> test_count 751.0000000 0.0000000000 751.0000000 0.0000000000
summary(nsp_res$error_fold)
#> mean sd median IQR
#> train.auroc 0.83743699 0.02048053 0.8333183 0.033646385
#> train.error 0.19173586 0.01438586 0.1967984 0.014375250
#> train.accuracy 0.80826414 0.01438586 0.8032016 0.014375250
#> train.sensitivity 0.88544232 0.03107595 0.8810630 0.039594807
#> train.specificity 0.65315955 0.07978799 0.6303327 0.123423546
#> train.fpr70 0.21096473 0.03745328 0.2102482 0.039804792
#> train.fpr80 0.26678331 0.04197401 0.2696150 0.055188199
#> train.fpr90 0.41645165 0.05634835 0.4302107 0.027201786
#> train.tpr80 0.67328390 0.09501731 0.7081574 0.136802093
#> train.tpr90 0.42606490 0.02625749 0.4288201 0.038100185
#> train.tpr95 0.24519417 0.10272755 0.2306974 0.135858369
#> train.events 333.33333333 6.47044563 334.0000000 5.750000000
#> train.count 500.66666667 0.51639778 501.0000000 0.750000000
#> test.auroc 0.59363657 0.03339583 0.5821419 0.002435316
#> test.error 0.37417530 0.02708388 0.3712590 0.010470120
#> test.accuracy 0.62582470 0.02708388 0.6287410 0.010470120
#> test.sensitivity 0.74203609 0.03728947 0.7396495 0.067245950
#> test.specificity 0.39550360 0.06362015 0.4024297 0.088476560
#> test.fpr70 0.56470600 0.04071028 0.5440798 0.046597213
#> test.fpr80 0.72560279 0.05383697 0.7249146 0.046820219
#> test.fpr90 0.85173443 0.03938630 0.8563455 0.020751291
#> test.tpr80 0.23524330 0.11922537 0.2271224 0.108842594
#> test.tpr90 0.12384858 0.07164059 0.1309382 0.038435952
#> test.tpr95 0.06825989 0.07977265 0.0511169 0.059644324
#> test.events 166.66666667 6.47044563 166.0000000 5.750000000
#> test.count 250.33333333 0.51639778 250.0000000 0.750000000
#> distance -1.00000000 0.00000000 -1.0000000 0.000000000
summary(nsp_res$represampling)
#> $`1`
#> n.train n.test
#> 1 500 251
#> 2 501 250
#> 3 501 250
#>
#> $`2`
#> n.train n.test
#> 1 501 250
#> 2 501 250
#> 3 500 251
#>
# plot(nsp_res$represampling, ecuador)
### Spatial cross-validation:
sp_res <- sperrorest(
data = ecuador, formula = fo,
model_fun = rpart,
model_args = list(control = ctrl),
pred_fun = mypred_part,
progress = TRUE,
smp_fun = partition_kmeans,
smp_args = list(repetition = 1:2, nfold = 3)
)
#> Thu Jan 30 04:22:55 2025 Repetition 1
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 1
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 2
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 3
#> Thu Jan 30 04:22:55 2025 Repetition 2
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 1
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 2
#> Thu Jan 30 04:22:55 2025 Repetition - Fold 3
summary(sp_res$error_rep)
#> mean sd median IQR
#> train_auroc 0.8472530 0.017474834 0.8472530 0.012356574
#> train_error 0.1820905 0.002353884 0.1820905 0.001664447
#> train_accuracy 0.8179095 0.002353884 0.8179095 0.001664447
#> train_sensitivity 0.8905000 0.009192388 0.8905000 0.006500000
#> train_specificity 0.6733068 0.011268634 0.6733068 0.007968127
#> train_fpr70 0.1842629 0.029580164 0.1842629 0.020916335
#> train_fpr80 0.2370518 0.011268634 0.2370518 0.007968127
#> train_fpr90 0.3555777 0.012677213 0.3555777 0.008964143
#> train_tpr80 0.7360000 0.053740115 0.7360000 0.038000000
#> train_tpr90 0.4845000 0.067175144 0.4845000 0.047500000
#> train_tpr95 0.2660000 0.024041631 0.2660000 0.017000000
#> train_events 1000.0000000 0.000000000 1000.0000000 0.000000000
#> train_count 1502.0000000 0.000000000 1502.0000000 0.000000000
#> test_auroc 0.5032968 0.024889595 0.5032968 0.017599602
#> test_error 0.4727031 0.028246609 0.4727031 0.019973369
#> test_accuracy 0.5272969 0.028246609 0.5272969 0.019973369
#> test_sensitivity 0.5470000 0.032526912 0.5470000 0.023000000
#> test_specificity 0.4880478 0.019720109 0.4880478 0.013944223
#> test_fpr70 0.7529880 0.061977487 0.7529880 0.043824701
#> test_fpr80 0.8884462 0.056343170 0.8884462 0.039840637
#> test_fpr90 0.9282869 0.000000000 0.9282869 0.000000000
#> test_tpr80 0.1610000 0.004242641 0.1610000 0.003000000
#> test_tpr90 0.0990000 0.007071068 0.0990000 0.005000000
#> test_tpr95 0.0650000 0.018384776 0.0650000 0.013000000
#> test_events 500.0000000 0.000000000 500.0000000 0.000000000
#> test_count 751.0000000 0.000000000 751.0000000 0.000000000
summary(sp_res$error_fold)
#> mean sd median IQR
#> train.auroc 0.84515941 0.01905839 0.84534979 0.028026750
#> train.error 0.18240710 0.01060702 0.18304622 0.009904172
#> train.accuracy 0.81759290 0.01060702 0.81695378 0.009904172
#> train.sensitivity 0.88941674 0.01990945 0.88819737 0.018551212
#> train.specificity 0.67338381 0.03405612 0.67399773 0.033408639
#> train.fpr70 0.19001989 0.02696925 0.18629168 0.035076259
#> train.fpr80 0.26077121 0.03429704 0.25571965 0.040338724
#> train.fpr90 0.38765695 0.03869441 0.38784530 0.057629711
#> train.tpr80 0.69871754 0.06961726 0.71852900 0.077197624
#> train.tpr90 0.38222915 0.12415524 0.37383597 0.127049184
#> train.tpr95 0.24811503 0.08804794 0.21752928 0.118033864
#> train.events 333.33333333 44.88503834 326.50000000 50.500000000
#> train.count 500.66666667 52.87217290 490.00000000 65.500000000
#> test.auroc 0.52865230 0.04745692 0.53136288 0.078210920
#> test.error 0.46155050 0.07803414 0.43884387 0.080997314
#> test.accuracy 0.53844950 0.07803414 0.56115613 0.080997314
#> test.sensitivity 0.58378570 0.19028544 0.64871054 0.191650843
#> test.specificity 0.47751069 0.16408495 0.45386905 0.161617488
#> test.fpr70 0.74420320 0.12039194 0.75586762 0.182721973
#> test.fpr80 0.84903452 0.12630703 0.87089783 0.074518397
#> test.fpr90 0.96263035 0.03429537 0.95823650 0.044154776
#> test.tpr80 0.15410646 0.07515119 0.15269750 0.066471765
#> test.tpr90 0.10004200 0.06560359 0.12034188 0.094070539
#> test.tpr95 0.04934735 0.07259254 0.01269036 0.075709340
#> test.events 166.66666667 44.88503834 173.50000000 50.500000000
#> test.count 250.33333333 52.87217290 261.00000000 65.500000000
#> distance -1.00000000 0.00000000 -1.00000000 0.000000000
summary(sp_res$represampling)
#> $`1`
#> n.train n.test
#> 1 498 253
#> 2 573 178
#> 3 431 320
#>
#> $`2`
#> n.train n.test
#> 1 482 269
#> 2 469 282
#> 3 551 200
#>
# plot(sp_res$represampling, ecuador)
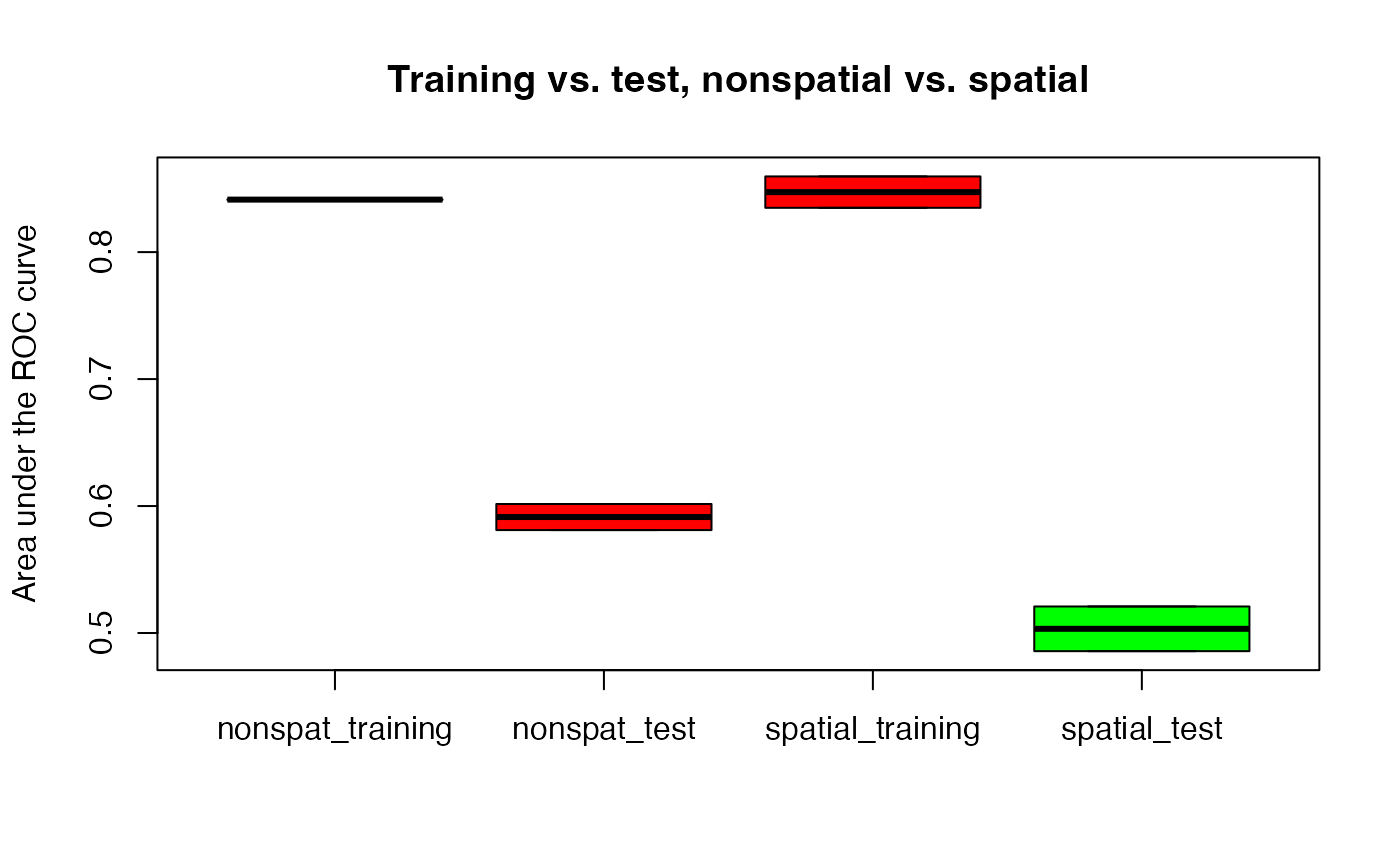
smry <- data.frame(
nonspat_training = unlist(summary(nsp_res$error_rep,
level = 1
)$train_auroc),
nonspat_test = unlist(summary(nsp_res$error_rep,
level = 1
)$test_auroc),
spatial_training = unlist(summary(sp_res$error_rep,
level = 1
)$train_auroc),
spatial_test = unlist(summary(sp_res$error_rep,
level = 1
)$test_auroc)
)
boxplot(smry,
col = c("red", "red", "red", "green"),
main = "Training vs. test, nonspatial vs. spatial",
ylab = "Area under the ROC curve"
)